DiRE identification of DIstant Regulatory Elements of co-regulated genes
OVERVIEW
DIRE.DCODE.ORG TRAFFIC
Date Range
Date Range
Date Range
LINKS TO WEBSITE
Bitnos is a free and collaborative Online Operating System that provides you all the best biomedical free web applications. Surf the website using the left sidebar and click on the star symbol. To insert here your favourite web applications. Physicochemical analysis of the primers. Protein structure and function prediction.
CREME has been retired and was replaced by DiRE. First, thank you all Creme users! Your interest in Creme has kept this service alive for over 4 years. Unfortunately, Creme has become outdated. It was operating on a very old version of the human genome. It was using outdated version of the TRANSFAC database. It was limited to the promoter regions of RefSeq genes only. Finally, we decided to retire the service.
Combining multiple methods developed by the community to predict causal noncoding variants. Prediction of regulatory motifs and motif combinations in a set of co-functional enhancers. Identification of proximal and Di. Prediction of synonymous regulatory elements in vertebrate genomes. Evolutionary conservation of multiple genomes. Identification and sequence analysis of regulatory elements.
We have recently modified ECR Browser alignments by including repetitive elements. All ECRbase ECRs correspond to new ECR Browser alignments now. Additionally, individual ECRs have been annotated according to their relationship to RefSeq genes and repetitive elements. And Transcription Factor Binding Sites in Vertebrate Genomes. Of RefSeq and UCSC known genes.
Zoom out 3x, i. Shift to the right,. Shift to the left, l. Flip the plot, g. Genome selection window, p. Reset parameters to defaults, f. Refresh the page, a. Main gene annotation, z.
Return to previously processed request.
Performs local multiple DNA sequence alignments of finished and draft-quality sequences. It identifies transcription factor binding sites evolutionarily conserved across multiple species. Return to previously submitted request.
Identifies transcription factor binding sites conserved across multiple species. Multiple sequence alignments of FINISHED sequences can be automatically submitted to multiTF from the results web page. Automatically forwards genome alignment data to Mulan for the subsequent multiTF post-processing. Return to previously submitted request.
Finding potential regulatory elements in noncoding regions of the human genome is a challenging problem. Analyzing novel sequences for the presence of known transcription factor binding sites or their weight matrices produces a huge number of false positive predictions that are randomly and uniformily distributed. If you have two sequence files.
Bps, but not more than. Fix the order of TFBS. Return to previously submitted request.
WHAT DOES DIRE.DCODE.ORG LOOK LIKE?
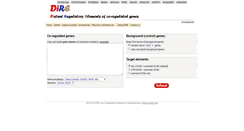
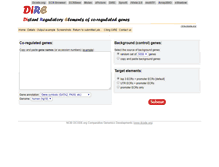
DIRE.DCODE.ORG SERVER
WEBSITE ICON

SERVER SOFTWARE
We discovered that dire.dcode.org is implementing the Apache server.SITE TITLE
DiRE identification of DIstant Regulatory Elements of co-regulated genesDESCRIPTION
Return to submitted job. Copy and paste gene names. Or accession numbers example. Gene symbols GATA2, PAX6, etc. GenBank mRNA AK, NM , BC, X, etc. Nucleotide RefSeq NM . Protein RefSeq NP . UCSC Known Genes uc000aaa.1. Select the source of background genes. Copy and paste background genes. Top 3 ECRs promoter ECRs default. UTR ECRs promoter ECRs. NCBI DCODE.org Comparative Genomics Developments www.dcode.org.PARSED CONTENT
The domain had the following on the web page, "Copy and paste gene names." I observed that the web site said " Or accession numbers example." They also stated " Gene symbols GATA2, PAX6, etc. GenBank mRNA AK, NM , BC, X, etc. Select the source of background genes. Copy and paste background genes. Top 3 ECRs promoter ECRs default. org Comparative Genomics Developments www."ANALYZE MORE BUSINESSES
Holsteinische Straße 4, 10717 Berlin. Wir freuen uns auf Ihren Besuch. Als AGFEO-Premium-Partner blicken wir auf eine 20-jährige Erfahrung im Bereich kleiner und mittlerer Telekommunikationslösungen zurück. Unser Schwerpunkt liegt hierbei auf der Verknüpfung von Apple-Systemen mit den vielfältigen Möglichkeiten von Agfeo-Telekommunikationssystemen.
Le style est vif , pressant , le lecteur.
Liedet, saunat, takat. Työpöydät, toimistopöydät ja ATK-pöydät. Hyllyt, lipastot, kaapit.
Neos Marmaras beach is only 250 meters on foot from the DIRE Hotel. Hotel Apartments Entertainment - Neos Marmaras - Halkidiki, Greece. DIRE is located in the heart of Neos Marmaras opposite the main church of the village; you have easy access to any kind of shop you want from bakery and grocery to bars and traditional taverns.
Nos valeurs et principes de formation.